Dr. Minggao Liang is a recent PhD graduate from Michael Wilson's lab at the Hospital for Sick Children and the University of Toronto, where his research focused on elucidating the cis-regulatory consequences of genetic variation in the context of evolution and human disease. Learn more about Minggao.
What are you working on?
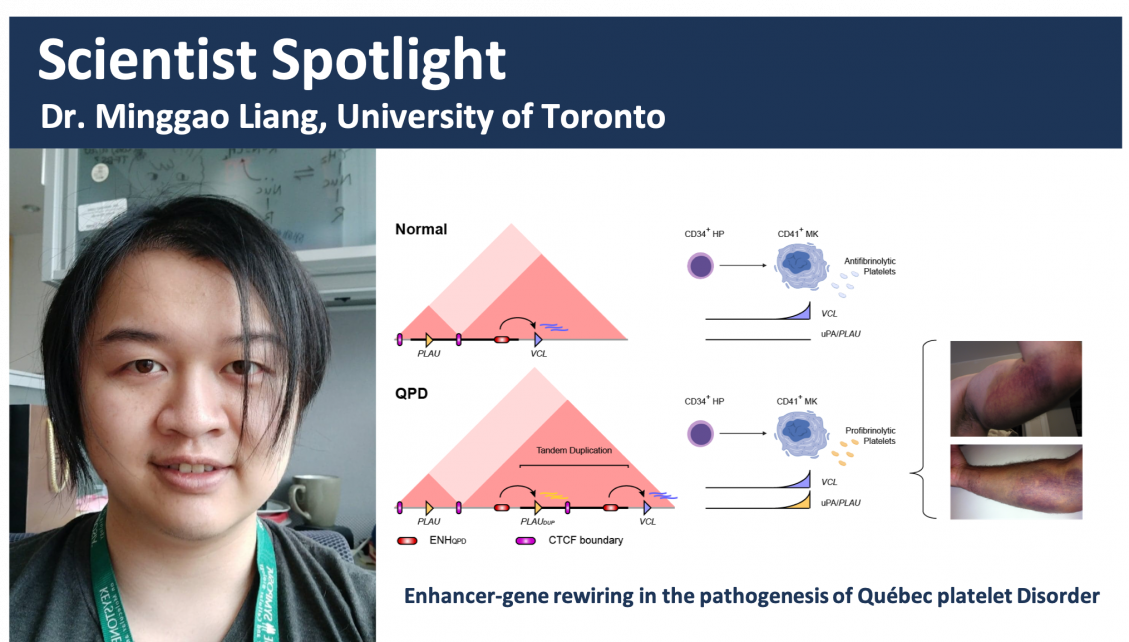
The accurate control of when and where genes are expressed is achieved in part by the folding of the genome into topologically associated domains (TADs) and subTADs. TADs demarcate physically interacting DNA domains in which enhancers can contact receptive gene promoters within the same TAD but are restricted from accessing gene promoters across TAD boundaries. Mutations that disrupt TAD organization can result in pathogenic dysregulation of gene expression through a mechanism of ‘enhancer hijacking’ and such mutations are increasingly being recognized in the etiology of human genetic disorders and cancers.
Quebec platelet disorder (QPD) is an autosomal dominant bleeding disorder with a unique, gain-of-function defect in fibrinolysis. The hallmark feature of QPD is a >100-fold overexpression of the profibrinolytic enzyme, PLAU, in platelets and megakaryocytes. The causative mutation is a 78-kb tandem duplication of PLAU, but how this duplication causes megakaryocyte-specific PLAU overexpression is unknown. To investigate the mechanism that causes QPD, we used epigenomic profiling, comparative genomics, and chromatin conformation capture approaches to study PLAU regulation in cultured megakaryocytes from participants with QPD and unaffected controls. The tandem duplication in QPD spans a subTAD boundary and led to ectopic interactions between PLAU and a conserved megakaryocyte enhancer found within the neighboring subTAD. Our results support a unique disease mechanism whereby the reorganization of subTAD genome architecture results in a dramatic, cell-type–specific blood disorder phenotype.
Where are you from? What do you miss about your hometown/country?
I was born in China but raised in Niagara and the GTA. I don’t remember much of my hometown, but what I miss most is the abundance of tasty late-night street food.
What city do you currently live in and what do you like most about this city?
I am currently based in Toronto. I love the diversity Toronto offers, especially when it comes to food!
What are some ways you detach from work/science/academia?
In my off time, I play and live-stream a strategic board game called GO. I used to play competitively before grad school and picked it up again during lockdown. I guess this counts as a special non-research talent, too.
Do you have a recommendation for a book, show, movie or documentary?
Check out “AlphaGo – The movie”. It’s a fantastic introduction to GO and how deep learning has transformed our approach to the game. Many of the paradigms are reflective of what machine learning is now doing for biology. Plus, it’s free to watch on youtube.
What made you decide to become a researcher?
A mix of environmental effects and cross-generational inheritance. Both my parents are in science and I had plenty of exposure growing up. There’s also the thrill that comes with discovery new things and understanding how things work.
If you weren't a researcher, what would you like to be/think you would be?
I’m not sure but probably a cook. Figuring out ways to prepare food shares many of the thrills as research and, as a bonus, is (usually) edible!
What sparked your interest in epigenetics?
I was fascinated by the question of how a single genome can give rise to hundreds of transcriptionally unique cell types in the body. Epigenetics is a big part of this puzzle.
If you could give your “first-year-PhD-self” one piece advice, what would it be?
Keep up a good work-life balance. Time and energy spent outside the lab are just as essential for healthy productivity as spent within.
Where would you like to see your research/field of interest end up in the future?
A key challenge right now is interpreting the cell type-specific effects of genetic variation on the epigenome. Something I’d like to see happen is being able to predict these effects computationally, in essence in silico epigenome editing, as a big step towards overcoming this challenge.